Chromatin remodelers are multidomain enzymatic
motor complexes that displace nucleosomes along DNA. A recent experimental breakthrough based
on cryo-EM allows now to understand how (some of the) remodellers work at the microscopic
level. Remodelers use the intrinsic mobility of nucleosomes that is based on twist defects in the
wrapped DNA, as described by us earlier (Kulic 2003).
They can be best understood as active Brownian dimers (Blossey 2019a)
that -- as they walk along the nucleosomal DNA -- create pairs of over- and under-twist defects,
see (Schiessel 2019) for a detailed commentary.

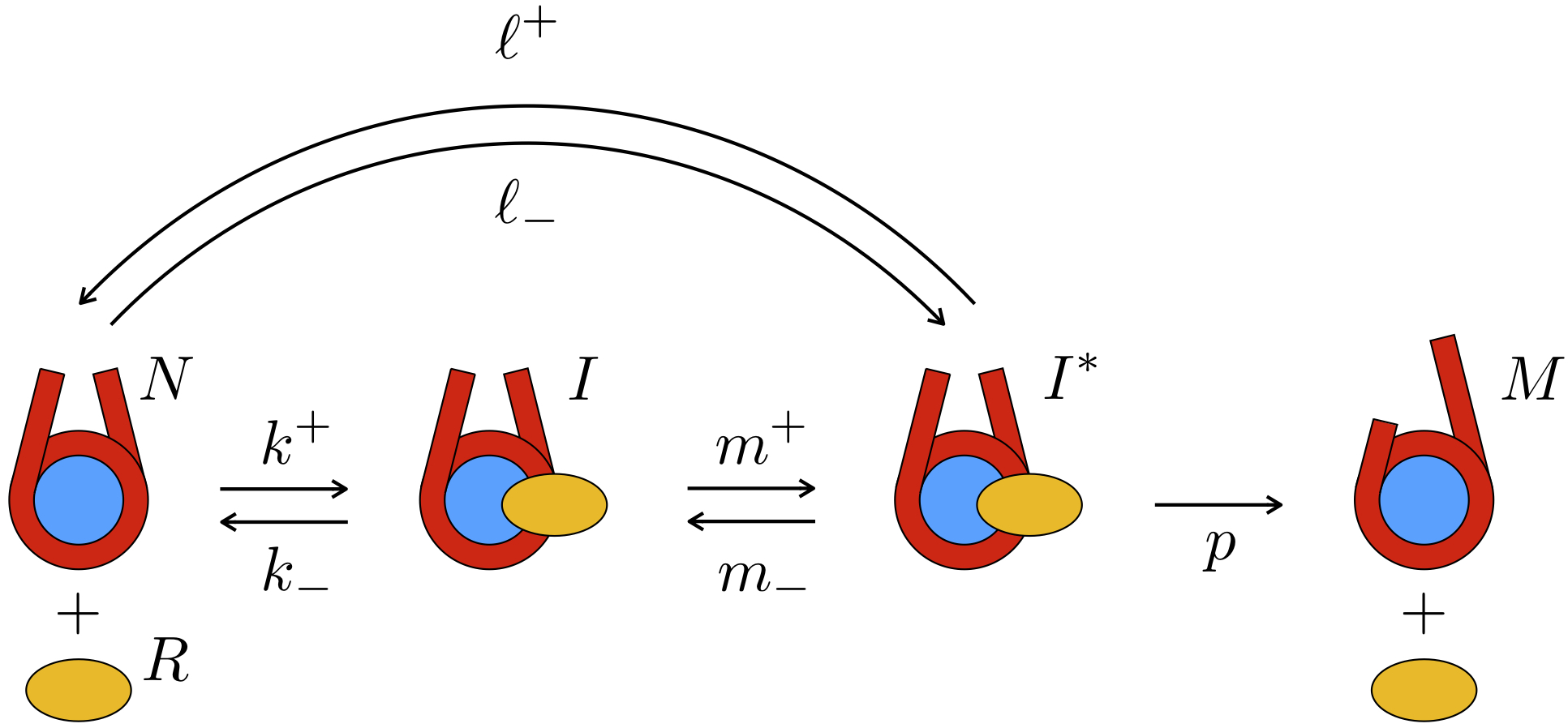
How do remodelers know on which nucleosomes they should act? In 2008 we put forward
a kinetic proofreading scheme that might explain how an ATP-consumption step of the remodeler
allows it to target specific nucleosomes (Blossey 2008).
Recent high-throughput data on nucleosome libraries assayed with remodelers are in accord with our earlier prediction of a
kinetic proofreading scenario (Blossey 2019b).
For details check out:
R. Blossey and H. Schiessel:
Chromatin remodelers as active Brownian dimers, J. Phys. A: Math. Theor.
52, 085601 (2019)
R. Blossey and H. Schiessel: Histone mark recognition controls nucleosome
translocation via a kinetic proofreading mechanism: Confronting theory and high-throughput experiments, Phys. Rev. E
99, 060401(R) (2019)
H. Schiessel: Nucleosomes as treadmills, Journal Club for Condensed Matter Physics, March 03 (2019)